In a study undertaken in more than 10,000 individuals, and published today in Nature Communications, scientists have shown that integrating information derived from different technologies to measure proteins can identify otherwise hidden links between proteins and human health and disease.
The international research team led by scientists from the Medical Research Council (MRC) Epidemiology Unit at the University of Cambridge provides a detailed view on how hundreds of plasma proteins are linked to hundreds of diseases and measures of health, showing that in several cases it might rather be the structure of a protein rather than its blood concentration that may mediate the effect.
Proteins are the essential functional units of the human body that are composed of amino acids and coded for by our genes. Naturally occurring sequence variation in the human genome can influence both the amount of a protein present and the function of the protein. Pharmaceutical companies and researchers now use this information to identify novel drug targets and treatment options. However, how we measure proteins shapes the insights we can gain, and up to now it has been unclear how this translates across thousands of proteins potentially linked to the whole spectrum of human health and disease.
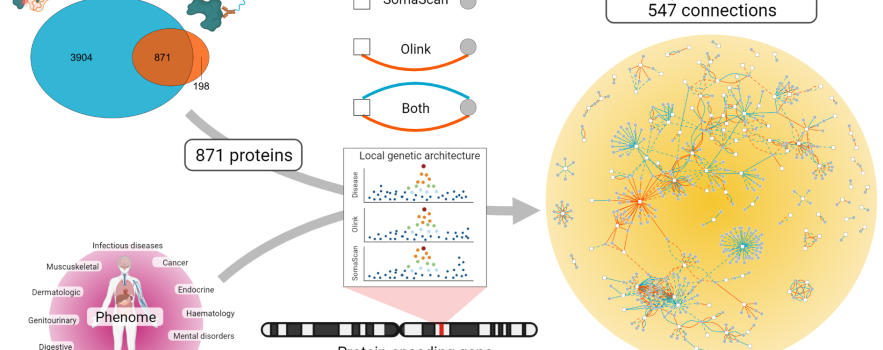
The researchers compared two commercially available proteomics technologies that each measure hundreds to thousands of proteins in blood samples simultaneously. The SomaScan v4 assay uses short, single-stranded DNA fragments, called aptamers, that fit to the structure of the protein like a key to a lock, whereas the Olink proximity extension assay uses sophisticated antibodies similar to those used in clinical labs. Using these complementary technologies, they measured the abundance of 871 circulating proteins overlapping between both technologies in more than 10,000 Fenland Study participants. They combined this with genetic data to chart a map showing how proteins contribute to diseases and health more generally.
One third of the more than 500 associations between individual proteins and disease or measures of health were only seen with one technology, highlighting a huge synergistic value of integrating technologies that goes beyond simple verification of results.
Dr Maik Pietzner, lead author of the study and researcher at the MRC Epidemiology Unit and Berlin institute of Health (BIH) at Charité Universistätsmedizin, Berlin, said:
To understand how proteins contribute to diseases we have to go beyond measuring the amount in blood. It was remarkable to see how taking differences in the results between the two technologies and combining these with genetics enabled us to identify novel biology. For example, we show that the role of the protein GDF-15 in severe nausea in pregnancy and childhood obesity, possibly through altering taste perception, does not depend on whether there is more or less of it in the blood, but rather on a minor change in its shape introduced by a genetic variant of which one in four persons carries at least one copy.”
However, the researchers also identified examples in which the results of the two proteomic assays for the genetic relationship of a protein with a particular phenotype differed. For example, a protein called PILRA seems to be causally involved in Alzheimer’s pathology, but whereas the Olink assay suggests higher levels of PILRA would be beneficial in preventing Alzheimer’s disease, the SomaScan assay correctly identifies that the opposite holds true. The researchers attributed such effects to the differences in the affinity of the two technologies for what they called ‘proteoforms’ of PILRA. The term proteoform describes proteins that differ in their shape or function due to common genetic variation that either changes single amino acids in the protein sequence or leads to multiple changes in the protein sequence by altering the genetic template that is used to encode the protein.
On a more technical note, the authors observed that both assays agreed moderately to well on roughly half of the proteins tested. They found that technical factors explained poor agreement, but also that common genetic variations that change the shape of proteins has a strong influence, basically disturbing the lock-and-key model. Despite unexpectedly high variation between both assays, they show that two in three genetic variants related to proteins were consistent between both platforms.
Dr Claudia Langenberg, of the MRC Epidemiology Unit, Cambridge, and BIH at Charité Universitätsmedizin, Berlin, Germany and senior author on the paper concluded:
We are just about to start to make full use of the enormous potential of combining protein measurements in blood with genetic variation to identify novel biology and diseases. Our study – for the first time – provides systematic evidence that is not only important for how we measure proteins, but also shows that complimentary technologies can provide synergistic insights into disease mechanisms and the development of treatment strategies. We generated a benchmark for future studies that aim to combine different technologies to maximize sample size for future discoveries.”
Reference
- Maik Pietzner, Eleanor Wheeler, Julia Carrasco-Zanini, Nicola D. Kerrison, Erin Oerton, Mine Koprulu, Jian’an Luan, Aroon D. Hingorani, Steve A. Williams, Nicholas J. Wareham, Claudia Langenberg. Synergistic insights into human health from aptamer- and antibody-based 1 proteomic profiling. Nature Communications 24 November 2021. DOI:10.1038/s41467-021-27164-0.

 MRC Epidemiology Unit
MRC Epidemiology Unit